|
chr1_-_161193190
|
8.796
|
|
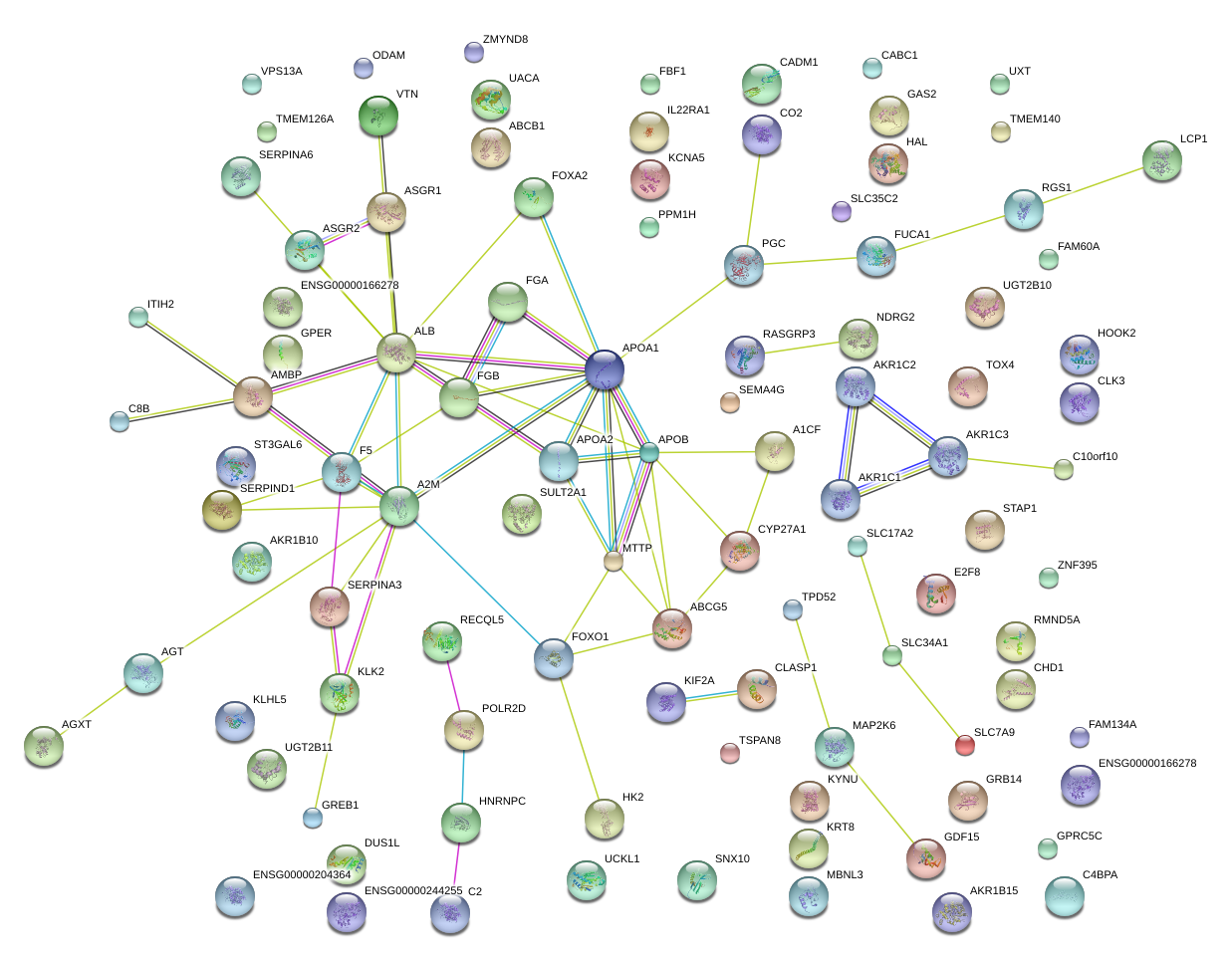
APOA2
|
apolipoprotein A-II
|
|
chr1_-_161193375
|
8.260
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr4_+_74269971
|
5.459
|
NM_000477
|
ALB
|
albumin
|
|
chr22_+_21128378
|
5.001
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr4_+_155484131
|
4.419
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr4_+_74270832
|
4.246
|
|
ALB
|
albumin
|
|
chr4_+_100495972
|
4.074
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr17_-_7082559
|
4.068
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr17_-_7082654
|
3.359
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr14_-_94789641
|
3.255
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr7_-_87342569
|
3.204
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr1_-_169555624
|
3.089
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr9_-_116840545
|
3.026
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr1_+_207277577
|
3.022
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr6_-_41715078
|
2.999
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr12_-_63328857
|
2.983
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr7_+_134832765
|
2.978
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr10_-_52645384
|
2.904
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_+_95078713
|
2.866
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr17_+_72427535
|
2.856
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chrX_-_131573638
|
2.732
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr10_-_52645430
|
2.553
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr9_-_116840460
|
2.511
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_-_69886112
|
2.316
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr1_-_169555710
|
2.282
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr20_-_45976626
|
2.195
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr9_-_116840551
|
2.188
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr17_-_26697293
|
2.170
|
|
VTN
|
vitronectin
|
|
chr6_+_31895201
|
2.104
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr19_+_18496966
|
2.102
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr12_-_53343607
|
2.056
|
|
KRT8
|
keratin 8
|
|
chr1_-_24469571
|
2.053
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr11_-_115375065
|
2.028
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr4_+_71062241
|
2.013
|
NM_017855
|
ODAM
|
odontogenic, ameloblast asssociated
|
|
chr20_-_22566092
|
1.993
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr17_-_26697310
|
1.978
|
|
VTN
|
vitronectin
|
|
chr13_-_46756326
|
1.974
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr17_-_26697224
|
1.972
|
|
VTN
|
vitronectin
|
|
chr11_+_22688156
|
1.962
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr13_-_41240607
|
1.953
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr4_-_155511838
|
1.937
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr11_-_116708334
|
1.927
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr1_+_227127993
|
1.926
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr19_-_48389514
|
1.916
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr2_+_33661392
|
1.916
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr1_+_227127959
|
1.902
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr10_+_5005453
|
1.893
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr10_-_5045967
|
1.864
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr10_+_5005601
|
1.844
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr13_-_46756295
|
1.841
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr2_+_11674241
|
1.833
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr2_-_165424993
|
1.825
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr12_-_71551564
|
1.754
|
|
TSPAN8
|
tetraspanin 8
|
|
chr3_-_12586962
|
1.749
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr1_+_227127999
|
1.729
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr12_-_96390002
|
1.672
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr17_-_26697358
|
1.646
|
NM_000638
|
VTN
|
vitronectin
|
|
chr2_+_219646448
|
1.631
|
NM_000784
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr8_-_80993009
|
1.612
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr1_-_24194760
|
1.575
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr6_-_25930838
|
1.553
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr2_+_75059781
|
1.553
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr3_+_98451537
|
1.544
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr17_-_7017971
|
1.534
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr10_+_102732285
|
1.522
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr10_+_7745341
|
1.522
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr2_+_220042948
|
1.493
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr9_-_116840604
|
1.437
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr7_+_134233848
|
1.415
|
NM_001080538
|
AKR1B15
|
aldo-keto reductase family 1, member B15
|
|
chr2_-_44065888
|
1.370
|
NM_022436
|
ABCG5
|
ATP-binding cassette, sub-family G (WHITE), member 5
|
|
chr10_+_5005685
|
1.363
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr4_+_39063851
|
1.355
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr12_-_31479038
|
1.343
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr20_-_62582478
|
1.329
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr5_-_98262217
|
1.322
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr2_+_86947387
|
1.319
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr14_-_21493122
|
1.303
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr2_+_143635225
|
1.287
|
|
KYNU
|
kynureninase
|
|
chr2_+_86947429
|
1.248
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr2_-_122159140
|
1.230
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr10_+_5136589
|
1.229
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr15_+_74907334
|
1.217
|
NM_001130028
|
CLK3
|
CDC-like kinase 3
|
|
chr10_+_5136556
|
1.216
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_+_192544856
|
1.208
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr12_-_9268522
|
1.199
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr2_+_143635194
|
1.194
|
NM_001032998
NM_001199241
NM_003937
|
KYNU
|
kynureninase
|
|
chr2_+_86947570
|
1.192
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr10_+_7745325
|
1.190
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr13_-_46756291
|
1.189
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr7_+_26400503
|
1.187
|
NM_001199837
|
SNX10
|
sorting nexin 10
|
|
chr9_+_79968344
|
1.173
|
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae)
|
|
chr17_+_67410825
|
1.155
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr7_+_45933096
|
1.133
|
|
|
|
|
chr7_+_1127722
|
1.128
|
NM_001098201
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr15_-_70994608
|
1.123
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr20_-_44993000
|
1.121
|
NM_015945
NM_173073
NM_173179
|
SLC35C2
|
solute carrier family 35, member C2
|
|
chr12_-_31479120
|
1.106
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr10_-_45474210
|
1.104
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr1_-_230850335
|
1.100
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_-_57431620
|
1.094
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr8_-_28243941
|
1.089
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr2_-_21266809
|
1.073
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr4_+_68424444
|
1.067
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr10_+_5136613
|
1.061
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr12_+_8332789
|
1.059
|
|
FAM66C
|
family with sequence similarity 66, member C
|
|
chr12_-_31479084
|
1.039
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr10_+_7745229
|
1.036
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr19_-_33360641
|
1.020
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr2_-_128615691
|
1.006
|
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D
|
|
chr7_-_140624280
|
1.002
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr3_-_183273407
|
1.002
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chrX_+_46940235
|
1.000
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr10_+_7745354
|
0.996
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr8_-_134309448
|
0.995
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr10_+_7745284
|
0.984
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr2_+_128176019
|
0.961
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr14_+_100563817
|
0.959
|
|
EVL
|
Enah/Vasp-like
|
|
chrX_-_48693934
|
0.952
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr7_-_25268057
|
0.950
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr15_-_65412175
|
0.947
|
|
PDCD7
|
programmed cell death 7
|
|
chr11_-_5526833
|
0.921
|
NM_001005567
|
OR51B5
HBE1
|
olfactory receptor, family 51, subfamily B, member 5
hemoglobin, epsilon 1
|
|
chr12_-_71551778
|
0.915
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chr1_+_220960038
|
0.906
|
NM_022746
|
MARC1
|
mitochondrial amidoxime reducing component 1
|
|
chr1_-_247054277
|
0.905
|
|
AHCTF1
|
AT hook containing transcription factor 1
|
|
chr1_-_231560789
|
0.895
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr6_-_76203256
|
0.895
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr11_-_65150107
|
0.881
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr1_+_227127937
|
0.861
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr9_-_116840673
|
0.855
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr4_+_74301932
|
0.855
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr14_-_21566728
|
0.848
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr18_-_52626636
|
0.836
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr14_-_20797532
|
0.826
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chrX_-_15288172
|
0.820
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr7_+_112063131
|
0.799
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr10_-_54531394
|
0.783
|
NM_000242
|
MBL2
|
mannose-binding lectin (protein C) 2, soluble
|
|
chr5_+_154092461
|
0.779
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr6_-_43027114
|
0.774
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr10_-_72648540
|
0.766
|
NM_000281
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha
|
|
chr4_-_89978117
|
0.765
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr5_-_42811959
|
0.763
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr2_-_27579277
|
0.761
|
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa
|
|
chr1_+_198608244
|
0.757
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_+_242127923
|
0.754
|
NM_001001666
NM_001001891
|
ANO7
|
anoctamin 7
|
|
chr1_+_236686738
|
0.753
|
NM_006499
NM_201543
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr5_-_131132642
|
0.738
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr21_+_34602304
|
0.735
|
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2
|
|
chr6_+_30130982
|
0.733
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr6_-_2176224
|
0.733
|
NM_001253846
|
GMDS
|
GDP-mannose 4,6-dehydratase
|
|
chr11_-_46615497
|
0.731
|
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr18_+_60382617
|
0.731
|
NM_194449
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr1_-_47695442
|
0.730
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr3_-_58522873
|
0.729
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr14_+_39736529
|
0.722
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr22_+_37309618
|
0.720
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr17_-_7018049
|
0.716
|
NM_001181
NM_001201352
NM_080912
NM_080913
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr17_-_29641068
|
0.711
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr8_-_110655418
|
0.710
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr12_-_103311380
|
0.706
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chrX_+_70315637
|
0.701
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr6_-_150057659
|
0.700
|
|
NUP43
|
nucleoporin 43kDa
|
|
chr2_-_65593866
|
0.684
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr10_-_118680978
|
0.681
|
|
KIAA1598
|
KIAA1598
|
|
chr14_+_102276279
|
0.668
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr4_-_151770611
|
0.666
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr5_+_70944968
|
0.653
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr13_+_36050885
|
0.649
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr8_+_107738239
|
0.647
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr3_-_11623829
|
0.644
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr12_-_9913415
|
0.643
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr17_-_29641087
|
0.633
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_-_49504337
|
0.632
|
|
LMBR1L
|
limb region 1 homolog (mouse)-like
|
|
chr18_-_74844710
|
0.625
|
|
MBP
|
myelin basic protein
|
|
chr1_+_50571963
|
0.624
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr21_+_34602205
|
0.620
|
NM_000874
NM_207584
NM_207585
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2
|
|
chr3_-_185826748
|
0.619
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr10_+_111985628
|
0.616
|
NM_005962
|
MXI1
|
MAX interactor 1
|
|
chr8_+_66933790
|
0.614
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr13_+_41885340
|
0.612
|
NM_001110798
NM_018527
NM_024561
|
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit
|
|
chr20_+_48552928
|
0.610
|
|
RNF114
|
ring finger protein 114
|
|
chr14_+_32798478
|
0.603
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr9_+_134000939
|
0.602
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr17_-_76124791
|
0.602
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr13_-_48987652
|
0.601
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr2_+_143886969
|
0.593
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr12_-_9268475
|
0.589
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr2_-_88926863
|
0.588
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr1_+_28764660
|
0.586
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr1_+_198608216
|
0.585
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr12_-_9268513
|
0.582
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr14_-_74551073
|
0.579
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr6_+_17600504
|
0.577
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr1_-_151414641
|
0.568
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr19_-_39826676
|
0.566
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr16_-_11375094
|
0.561
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr4_+_187187342
|
0.561
|
|
F11
|
coagulation factor XI
|
|
chr20_+_48552935
|
0.561
|
|
RNF114
|
ring finger protein 114
|
|
chr7_+_44646180
|
0.558
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr11_-_102826433
|
0.553
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr7_-_87230170
|
0.548
|
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|